Getting started with computational biology in cancer research
13 March 2019
I‘m Dr Ben Hall, Programme Leader at the research centre at MRC Cancer Unit (University of Cambridge). In my first guest blog, I’d like to discuss a new branch of modern biology known as computational biology, and how it’s relevant to OCR’s A Level Biology specification, particularly in the areas of genome sequencing and bioinformatics.
In this blog post I’ll share examples of network and testing models using the BioModelAnalyzer, as well as links to online activities and tutorials you can download and use.
We also produced a 3 minute film for you to watch about our work on cancer progression using computers.
What is computational biology?
Computational biology involves the development of computational simulations, mathematical modelling and bioinformatics. Using as a part of A Level practical work allows students to show competency in the use of ICT such as computer modelling to collect data.
Computational biology uses computers to support our understanding of biological processes. In research labs, this speeds up our analysis of experiments but also lets us make new predictions based on simulations.
Our team have developed a PAG10 exercise around a computational tool, the BioModelAnalyzer, which demonstrates how different proteins may drive cancer, as well as why not all drugs work equally well for all cancers.
To introduce new terms and ideas, we’ve also created a pre-activity worksheet that introduces cancer biology. Here we will highlight some of the tasks in the activity that can be used in your classes.
Exploring network models using the BioModelAnalyzer
 At heart of cancers are cells. Cancer cells are the descendants of healthy cells which have accumulated errors, resulting in changes in cell behaviour that drive the cancer forward.
At heart of cancers are cells. Cancer cells are the descendants of healthy cells which have accumulated errors, resulting in changes in cell behaviour that drive the cancer forward.
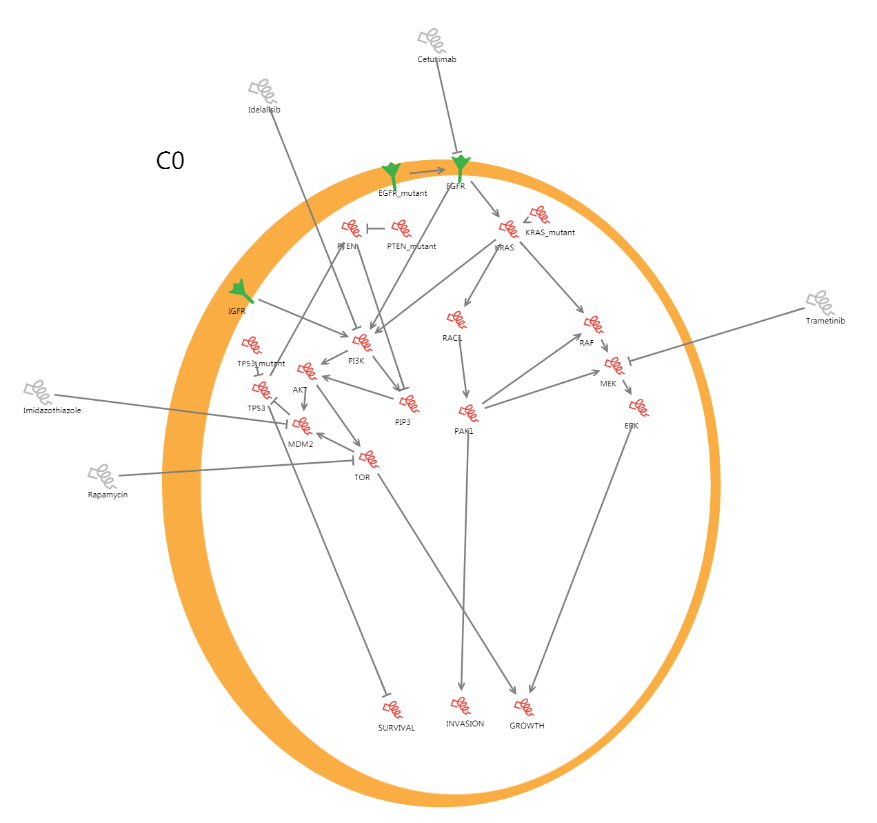
The starting point for understanding a cancer is to look at how the behaviour of the healthy cell is controlled. To do this we construct a network of the genes in the cell describing how this occurs.
The BioModelAnalyzer offers a canvas onto which you can add cells, genes, and proteins with ease by dragging and dropping. We have included one model with the practical that you can examine.
The cell is an orange circle, with proteins on the surface of the cell in green, whilst intracellular and extracellular molecules are represented by red and grey helices. If one molecule activates another it has a pointed arrow leading to it, whilst if it inhibits another it is linked with a flat headed arrow.
At the bottom of the cell we have included molecules that represent the overall behaviour of the cell. This is determined by the activities of molecules in the cell that link directly to it.
Testing models
A network is like a map of the cell. It can tell you about who is talking to who, but it cannot on its own say how the cell might change over time.
You might be interested in how it changes if one gene is lost, or if the cell receives a signal from a neighbouring cell. To do this you need to consider the state of the genes and how they change in response to their neighbours.
To understand how the cell behaves you need to define the states and specify rules that say how they update. In this model, every molecule can either be off (zero) or on (one). A simulation tells us how the cell changes over time from the initial starting state by repeatedly calculating the next states from the inputs for each variable.
Mutating cells and seeing how they change
If the model represents a healthy cell in a tissue, we can look for genes that are changed in cancer. For example, TP53 is known as a “tumour suppressor” as turning it off is thought to promote cancer.
Another protein, KRAS, is called an “oncogene” as mutations that make it permanently active promote cancer. We can trace how these changes propagate through the network to change the cell behaviour and ultimately promote cancer.
By clicking on molecules in BioModelAnalyzer we can turn them permanently off or on, in the same way a gene might be mutated in the progression of cancer. Turning the molecule permanently on or off may change other molecules in the network through its interactions. This activity propagates through the model ultimately leading to the changes in behaviour.
Download the resources
To conclude, these are the basics of what BioModelAnalyzer can show.
My team and I have created additional activities that cover PAG10 requirements in the field of computational methods in biology. You can access the resources at the following links:
biomodelanalyzer.org/Teachers
biomodelanalyzer.org/Students
As well as the worksheets, there is a tutorial that demonstrates how to do other activities, including working with larger networks and building your own from scratch.
When you use this practical activity please let me know how it goes. You can email me if you have any questions, need clarifications, or feedback at bh418@mrc-cu.cam.ac.uk.
Stay connected
If you have any questions for the science team at OCR please submit your comments below or email them directly with your questions to either science@ocr.org.uk.
You can also sign up to receive email updates about any of our subjects or follow us on Twitter at @OCR_Science.
About the author
Dr Ben Hall

Dr Hall is a computational biologist in the MRC Cancer Unit. He leads a program on “Modelling the decision processes of cancer”, where his team develop computational models of the earliest stages of cancer progression, with a particular focus on the use of applying new techniques from computer science. He holds a Royal Society University Research Fellowship, and work in his group is funded by the MRC, Microsoft Research, and the Royal Society.